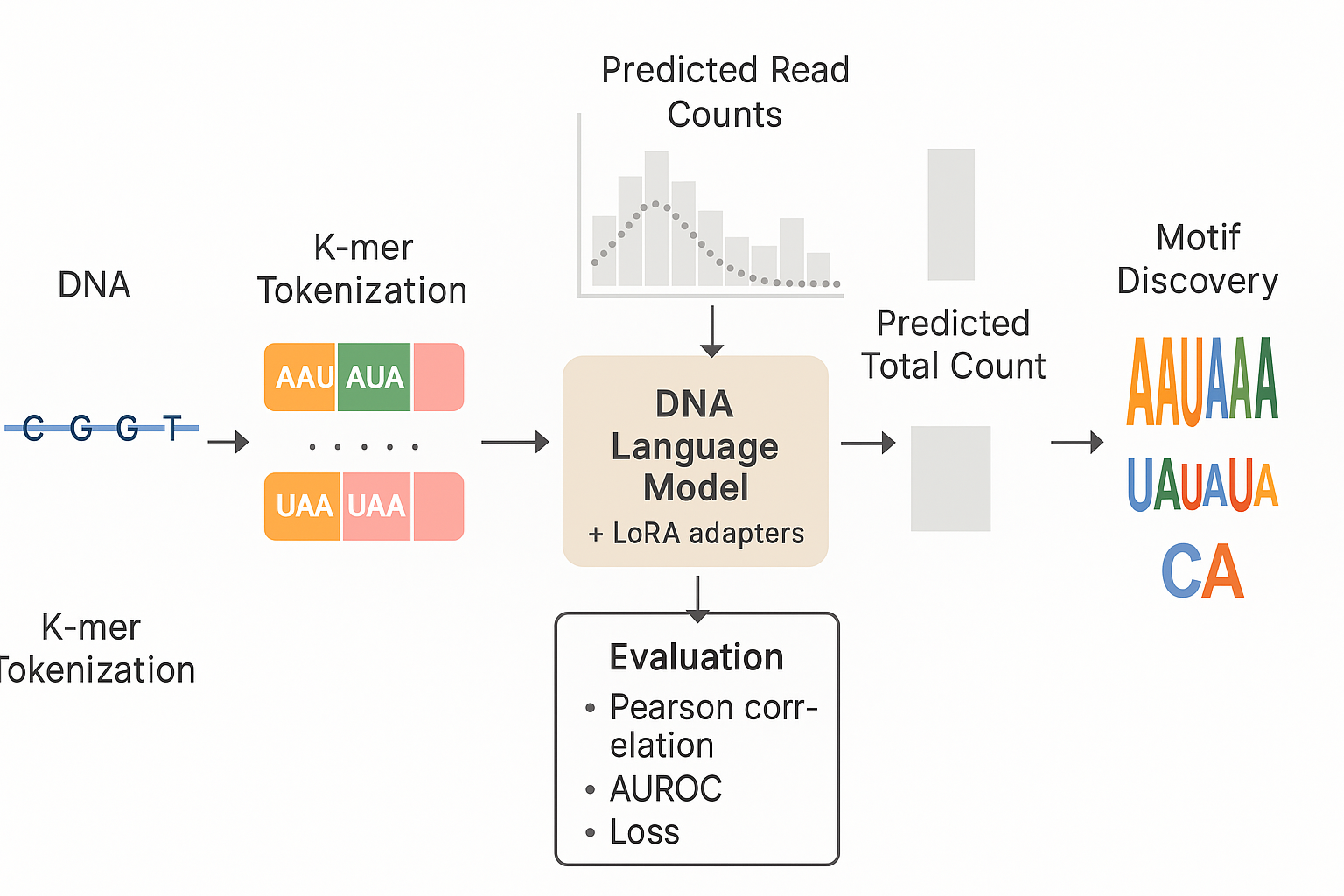
- We build an end-to-end ML pipeline for regulatory genomics: data prep, tokenizer/species tags, training (BPNet and SpeciesLM), hyperparameter sweeps, evaluation, and motif interpretation with DeepLIFT → TF-MoDISco.
- LoRA-tuned SpeciesLM/DNABERT outperforms BPNet and a frozen-LM on loss, Pearson correlation, and AUROC for poly(A) site prediction.
- The pipeline yields interpretable biology: recovery of AAUAAA/variants, UAUAUA, and CA at the cut site, with expected variability.
- Efficient and practical: adapter-based fine-tuning keeps compute and memory low while improving accuracy.
🔗 Shortcuts
📚 Abstract
The genome encodes regulatory signals beyond protein-coding regions. In particular, 3′-UTR polyadenylation relies on upstream/downstream sequence elements whose diversity complicates classical alignment-based analyses. We frame poly(A) site prediction as a sequence modeling task and evaluate:
- BPNet as a strong, interpretable CNN baseline, and
- a Species-aware DNA LM (DNABERT/SpeciesLM) with (i) a shallow 1D-conv prediction head or (ii) a BPNet-style head.
We fine-tune the LM using Low-Rank Adaptation (LoRA) to efficiently adapt encoder blocks. Our best model (SpeciesLM + LoRA) achieves lower validation/test loss, higher Pearson correlation, and better AUROC than both the BPNet baseline and a frozen-LM alternative. With TF-MoDISco, we validate that learned features align with literature-reported polyadenylation elements, while noting limitations in motif spacing/order recoverability from local attributions.
🚀 Highlights
- Models: BPNet baseline vs. SpeciesLM (DNABERT-style encoder, 12 layers, ~90M params).
- Fine-tuning: LoRA on encoder blocks → ~2.4M trainable params instead of full 90M.
- Heads: (a) shallow 1D-conv; (b) BPNet-like dilated conv stack.
- Interpretability: DeepLIFT → TF-MoDISco recovers AAUAAA/UAUAAA/AAAAAA, UAUAUA, and CA motifs.
- Takeaway: LoRA outperforms parameter-heavy alternatives while remaining compute-friendly.
🧪 Datasets & Preparation (Yeast focus)
- Regions centered on candidate poly(A) sites assembled from genomic counts + TIF-seq.
- Inputs standardized to 300 bp;
- BPNet: one-hot nucleotides.
- SpeciesLM: 6-mer tokenization + species token (“yeast”).
- Chromosome splits:
- Train: I–XIV, XVI
- Val: XV
- Test: VII
🧠 Models & Training
Baseline — BPNet
- Dilated CNN with residual connections.
- Loss = profile (Multinomial NLL) + counts (MSE).
Language Model — SpeciesLM (DNABERT-style)
- 12-layer encoder (~90M params), k-mer tokens, bidirectional self-attention.
- Heads:
- Shallow 1D-conv reduction (768 → 512 → 256 → 128 → prediction)
- BPNet-style dilated stack on top of LM embeddings
LoRA Fine-tuning
- Freeze base weights; train low-rank adapters inside attention/FFN layers.
- Benefits: small trainable footprint, stable optimization, lower memory.
📊 Results
Test-set performance (best checkpoints by val loss):
| Model | Pearson (Median) | Pearson (Mean) | AUPRC | AUROC | Test Loss |
|---|---|---|---|---|---|
| BPNet | 0.730 | 0.682 | 0.605 | 0.920 | 939.203 |
| SpeciesLM + LoRA | 0.809 | 0.739 | 0.640 | 0.931 | 711.484 |
| SpeciesLM (frozen) | 0.771 | 0.703 | 0.623 | 0.926 | 844.048 |
Key observation: LoRA delivers the strongest overall metrics and smooth optimization without full-model unfreezing.
🔍 Motif Discovery (TF-MoDISco)
- Poly(A) site signals: clear CA dinucleotide with downstream A-stretch and flanking T’s.
- Positioning elements: AAUAAA, UAUAAA, occasionally AAAAAA variants.
- Efficiency element (yeast): TA-rich patterns consistent with UAUAUA.
- Caveat: Local attributions limit conclusions about global spacing/order between motifs.
🧰 Reproducibility
Clone the repository and install dependencies:
git clone https://github.com/<your-org>/<your-repo>.git
cd <your-repo>1) Environment
# create and activate conda env
conda env create -f environment.yml
conda activate regulate-meIf using GPU, install a PyTorch build matching your CUDA version.
2) Data
# place your dataset here
mv saccharomyces_cerevisiae ./data
# one-time preprocessing
python scripts/preprocess_data.py3) Training (Hydra configs)
# BPNet baseline
python scripts/train_model.py --config-name config
# SpeciesLM + shallow conv head (LoRA on by default)
python scripts/train_model.py --config-name lora
# SpeciesLM + BPNet-style head
python scripts/train_model.py --config-name llm_bpnetToggle LoRA (example):
python scripts/train_model.py --config-name lora model.use_lora=False4) Sweeps (W&B)
wandb login
python scripts/run_sweep.py --config-name llm_bpnet🧑🤝🧑 Contributions
- MR — Biological research & interpretation; wrote polyadenylation background.
- PN — Model design/implementation/training; attribution & MoDISco pipeline; methods & results.
- HA — Results interpretation; model background; parts of discussion; feedback.
- SZ — Interpretation pipeline (DeepLIFT & TF-MoDISco) implementation.
🙌 Acknowledgements
This project was conducted in ML4RG (SS24) at TUM. We thank our instructors and peers for feedback and support.
