round(runif(5, 1, 2), 2)[1] 1.09 1.67 1.47 1.66 1.90Peter Nutter
Sunday, April 28, 2024
runif(n):p: cumulative distributionr: random variabled: probability distributionq: quantile| Distribution | R Tag | Parameters |
|---|---|---|
| Beta | beta | shape1, shape2 |
| Binomial | binom | size, prob |
| Chi-squared | chisq | df |
| Exponential | exp | rate |
| F | f | df1, df2 |
| Gamma | gamma | shape, rate or scale |
| Geometric | geom | prob |
| Lognormal | lnorm | meanlog, sdlog |
| Negative Binomial | nbinom | size, prob |
| Normal | norm | mean, sd |
| Poisson | pois | lambda |
| Student’s t | t | df |
| Uniform | unif | min, max |
Check:
Let \(X\) be a continuous random variable with CDF \(F_X\) (assume for simplicity \(F_X\) is strictly increasing).
\[ F_X(X) \sim U(0,1) \]
Proof: - CDF of \(U(0,1)\) is \(F_U(u) = u\) \[ P(F_X(X) \leq u) = P(X \leq F_X^{-1}(u)) = F_X(F_X^{-1}(u)) = u \]
Method: - Generate random uniform \(U \sim U(0,1)\). - Set \(X = F_X^{-1}(U)\).
Example: Generate from density \(3x^2\) for \(x\) in \([0, 1]\). - \(F_X = x^3\) - \(x = F_X^{-1}(u)\) - \(x = u^{1/3}\)
Code:
Algorithm:
Note: In practice, \(1 - u\) is often used to avoid issues with \(u = 0\).
Algorithm: - Generate random uniform \(U \sim U(0,1)\). - Set \(X = x_i\) if \(F_X(x_{i-1}) < U \leq F_X(x_i)\).
Example: \(X\) Bernoulli(p) - \(P(X=0) = 1-p\) - \(P(X=1) = p\) - \(F(0) = 1-p\) - \(F(1) = 1\) \[ X = I(1-p < U \leq 1) = I(1-p < U) \]
We discretize the quantile line in the proportion of the discrete probabilities.
Proof: \[ P(y|\text{accept}) = \frac{P(\text{accept}|Y) g(y)}{P(\text{accept})} = \frac{f(y) g(y) / M g(y)}{1/M} = f(y) \]
Example: Beta distribution - Generate 1000 samples from Beta(2,2): \[ f(x) = \frac{\Gamma(2+2)}{\Gamma(2) \Gamma(2)} x^{2-1} (1-x)^{2-1} = 6 x (1-x) \] - Support of Beta is \([0,1]\). - \(g\) is uniform and \(M = \frac{3}{2}\).
Algorithm: - Generate \(Y \sim U(0,1)\) - Generate \(U \sim U(0,1)\) - Accept \(Y\) if: \[ U \leq \frac{6 Y (1-Y)}{\frac{3}{2}} = 4 Y (1-Y) \]
::: {.cell}
::: {.cell-output-display} 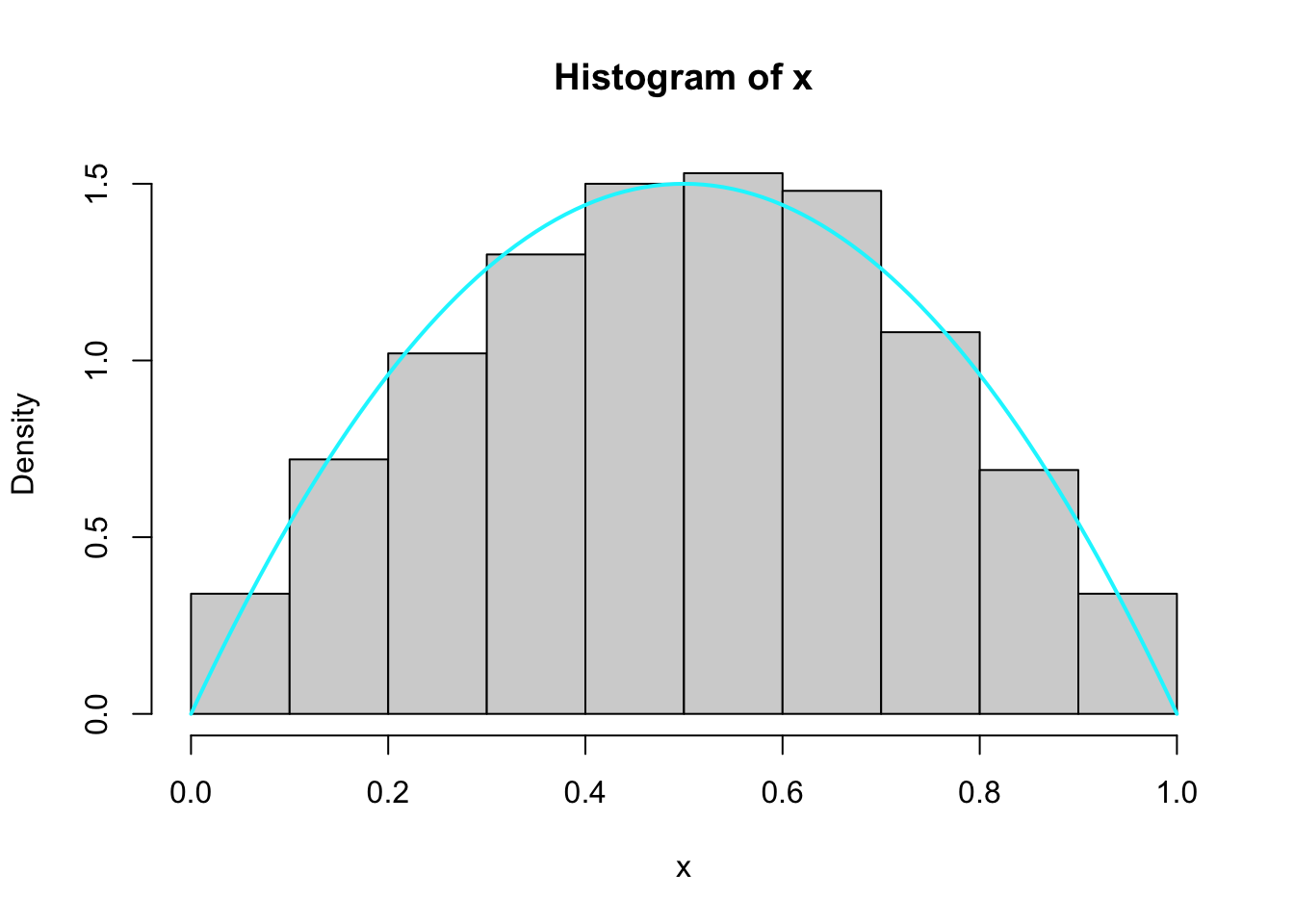 :::
:::
::: {.cell-output .cell-output-stdout}
[1] 1532::: :::
\[ \text{se}(x_q) = \sqrt{\frac{p(1-p)}{n f^2(x_q)}} \]
Agreement between 1 and 2 SE is considered good:
samples theoretical diff
10% 0.1944411 0.1958001 -0.15162513
20% 0.2905545 0.2871407 0.21974679
30% 0.3742096 0.3632575 0.54457511
40% 0.4435312 0.4329311 0.46451623
50% 0.5107799 0.5000000 0.45451925
60% 0.5721829 0.5670689 0.22410331
70% 0.6347660 0.6367425 -0.09827994
80% 0.7093491 0.7128593 -0.22595231
90% 0.8038251 0.8041999 -0.04181931Relationship between common distributions:
\(Z \sim N(0,1)\) then \(Z^2 \sim \chi^2_1\)
\(U \sim \chi^2_m\) and \(V \sim \chi^2_n\) independent then \(F = \frac{U/m}{V/n} \sim F_{mn}\)
\(T\) distribution:
Example: simulate Beta from Gammas - Relationship does not depend on \(\lambda\). - Generate Beta(3,2):
n <- 1000
beta <- 2
alpha <- 3
u <- rgamma(n, shape = alpha, rate = 1)
v <- rgamma(n, shape = beta, rate = 1)
x <- u / (u + v)
p <- seq(.1, .9, .1)
samples <- quantile(x, p)
theoretical <- qbeta(p, alpha, beta)
plot(samples, theoretical, las = 1, pch = 21, bg = "red")
abline(0, 1, lwd = 2, col = "lightblue")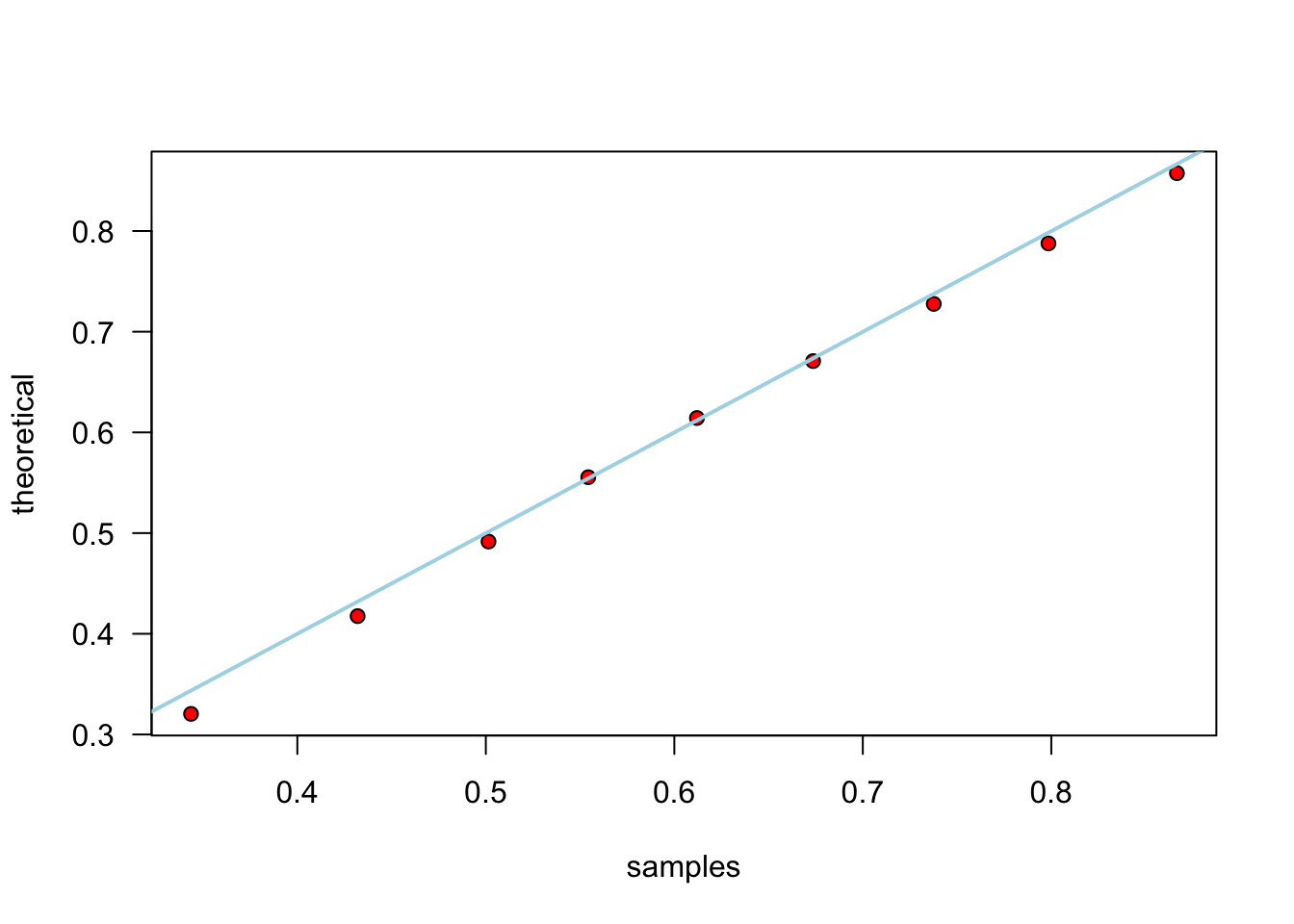
Examples:
Example: generate \(n\) \(\chi^2_{v}\):
Compare theoretical and empirical quantiles:
We show this using a transformation method:
Continuous: \[ X \sim f = \int_{-\infty}^{\infty} f_{X|Y} f_Y dy \] - \(f_{X|Y}\) family of distributions indexed by \(y\) weighted by \(f_Y\).
Example: Mixture of normals - \(f_X = \frac{1}{2} N(0,1) + \frac{1}{2} N(2,1)\)
Algorithm:
Implementation:
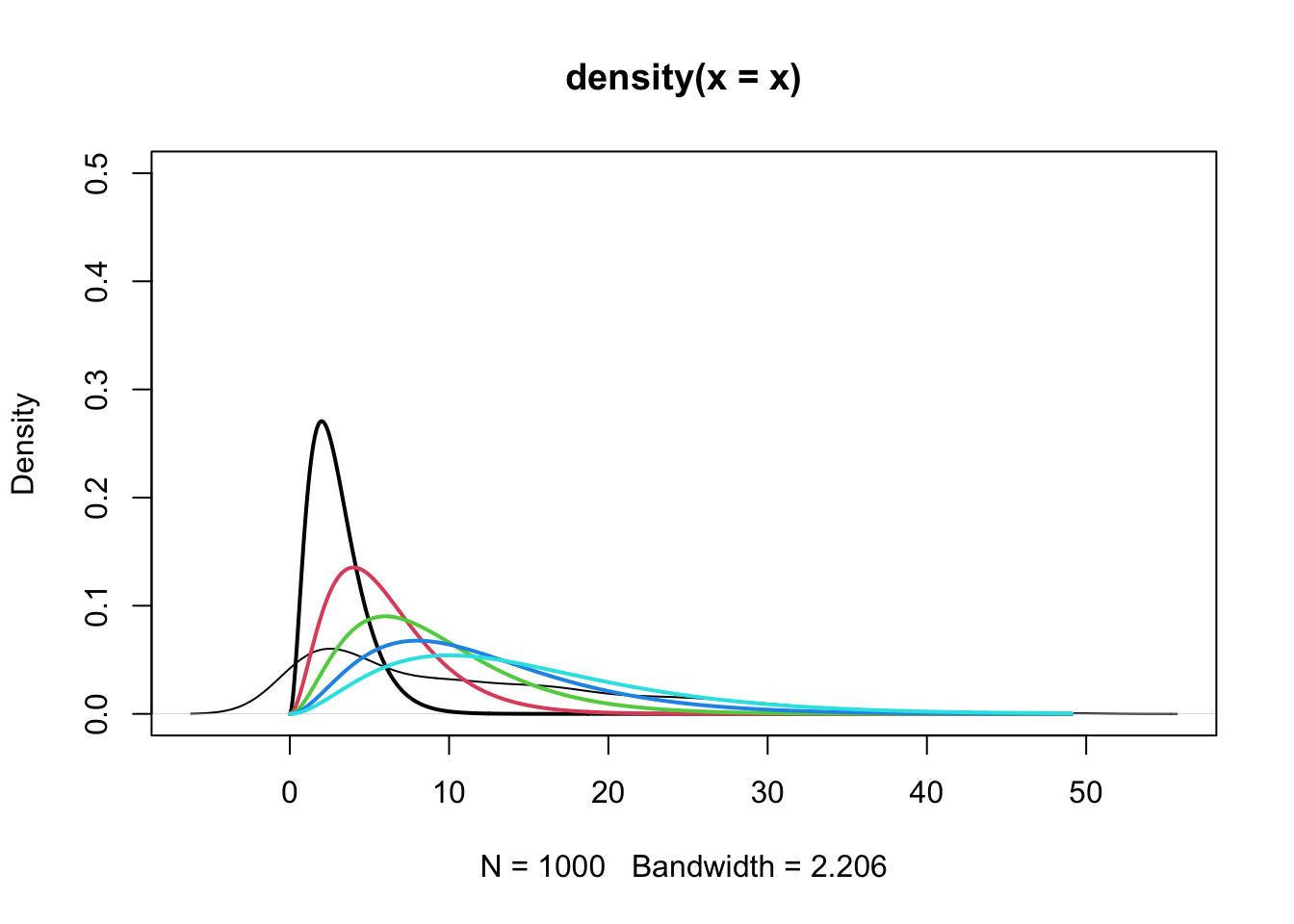
Example: Mixture of gammas
Mean of \(f_{X_i}\): \(3i\)
Mean of \(f_X\): \[ \sum_{i=1}^{5} \frac{i}{15} \cdot 3i = 11 \]
::: {.cell}
::: {.cell-output .cell-output-stdout}
[1] 11::: :::
Generate 1000 samples:
n <- 1000
pi <- 1:5 / 15
rates <- 1 / (1:5)
mat <- matrix(rgamma(n * 5, shape = 3, rate = 1 / (1:5)), n, 5)
x <- rowSums(mat * pi)
plot(density(x), ylim = c(0, 0.5), col = "black")
# overlay plots of the 5 mixture densities of the gammas
y <- seq(0, max(x), length.out = 1000)
for (i in 1:5) {
lines(y, dgamma(y, shape = 3, rate = rates[i]), col = i, lwd = 2)
}
[1] 11.1973Negative binomial is a Poisson-Gamma mixture:
Discrete distribution: number of failures before \(r\) successes in a sequence of independent Bernoulli trials with success probability \(p\): \[ f_X = \binom{x+r-1}{x} p^r (1-p)^x \]
::: {.cell}
::: {.cell-output-display} 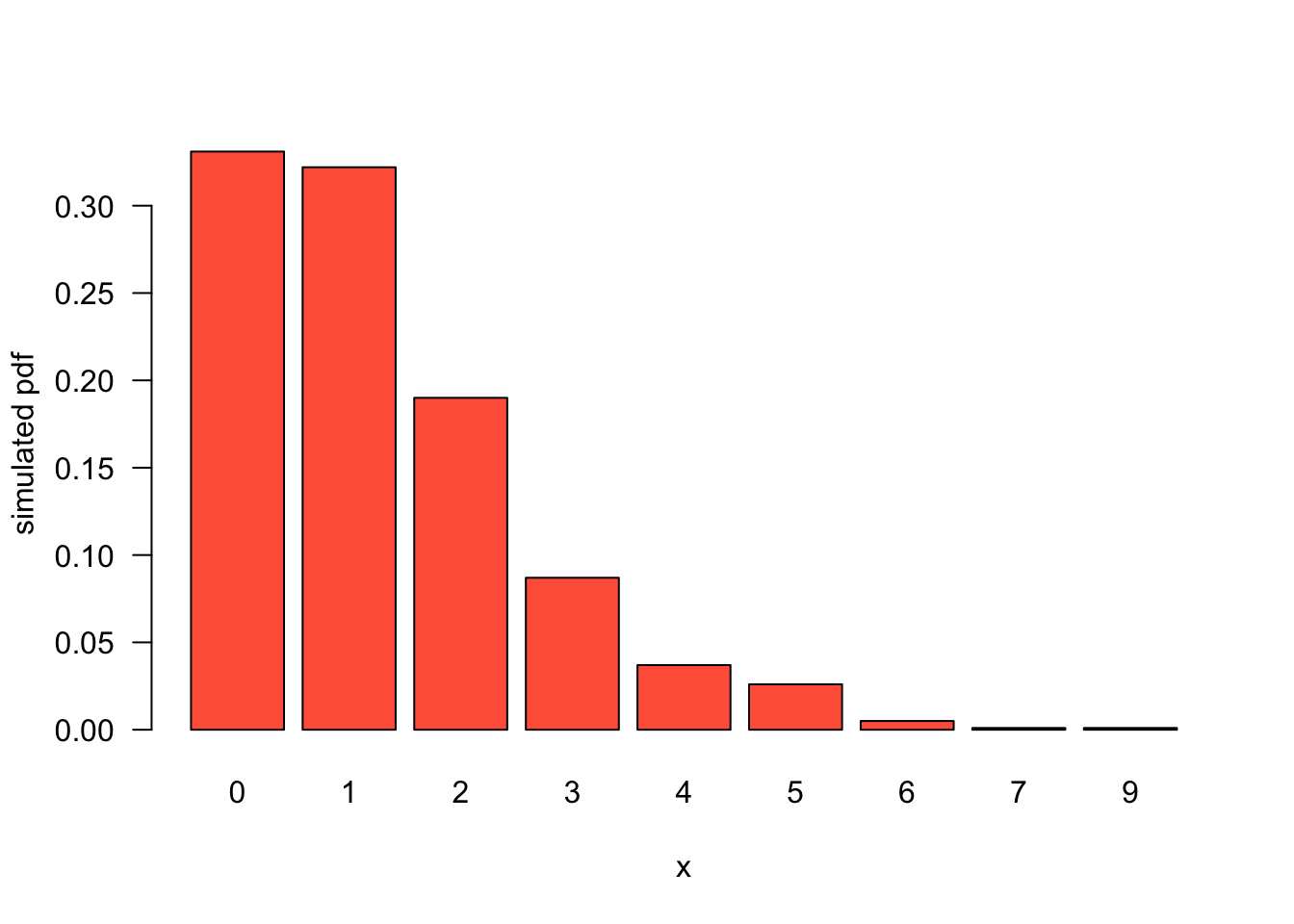 ::: :::
::: :::
Comparing the theoretical and empirical quantiles for discrete distributions does not work well because the theoretical quantiles are not unique. - Standard error of ECDF is a better way to compare: \[ \text{se}(F_x) = \sqrt{\frac{F_n (1-F_n)}{n}} \]
::: {.cell}
::: {.cell-output .cell-output-stdout}
0 1 2 3 4 5 6 7 9
unique_x 0.000 1.000 2.000 3.000 4.000 5.000 6.000 7.000 9.000
sim 0.331 0.322 0.190 0.087 0.037 0.026 0.005 0.001 0.001
truth 0.316 0.316 0.198 0.099 0.043 0.017 0.006 0.002 0.000
diff 0.015 0.006 -0.008 -0.012 -0.006 0.009 -0.001 -0.001 0.001
two_se 0.029 0.029 0.025 0.019 0.013 0.008 0.005 0.003 0.001::: :::
mvnorm in the MASS package
rmvnorm in the mvtnorm package
We can generate them from \(N(0,1)\).
\[ X \sim N(\mu, \sigma^2) \] \[ X = \sigma Z + \mu \]
Proof it works by the density transformation formula:
We extend this scale and shift result to the multivariate case.
Multivariate normal generator:
Algorithm:
chol function:
n <- 100000
p <- 3
mat <- matrix(rnorm(n * p), n, p)
epsilon <- matrix(rnorm(p * p), p, p)
epsilon <- t(epsilon) %*% epsilon
Q <- chol(epsilon)
m <- rnorm(p)
mu <- matrix(m, n, p, byrow = TRUE)
x <- mat %*% Q + mu
# shape
# x[1,] # corresponds to a rv with mean mu and covariance epsilon the first one
mean <- apply(x, 2, mean)
round(cov(x), 1) [,1] [,2] [,3]
[1,] 5.9 1.6 -2.7
[2,] 1.6 2.5 0.4
[3,] -2.7 0.4 2.4 [,1] [,2] [,3]
[1,] 5.9 1.7 -2.7
[2,] 1.7 2.6 0.4
[3,] -2.7 0.4 2.5The chi-square is often used to model sample variances \(s^2\):
The Wishart distribution extends this to distributions for positive definite symmetric sample covariance matrices \(X \in \mathbb{R}^{p \times p}\) estimating a multivariate Normal variance \(V\):
\[ f(X) = \frac{|X|^{\frac{n-p-1}{2}}}{2^{\frac{np}{2}} |V|^{\frac{n}{2}} \Gamma_p\left(\frac{n}{2}\right)} \exp\left(-\frac{\text{tr}(V^{-1}X)}{2}\right) \] with \(\Gamma_p(\cdot)\) the multivariate gamma function and \(|\cdot|\) the determinant. This density is called \(W_p(V, n)\).
Generating from the Wishart distribution:
Recall from earlier:
Generalize to:
n_samples <- 1000
m <- 10
n <- 10
# Function to generate F distribution samples
generate_f <- function(n_samples, m, n) {
# Generate chi-squared random variables with m degrees of freedom
chim <- rchisq(n_samples, m)
chim <- chim / m
# Generate chi-squared random variables with n degrees of freedom
chin <- rchisq(n_samples, n)
chin <- chin / n
# F distribution is the ratio of two scaled chi-squared variables
f <- chim / chin
return(f)
}
# Generate F distribution samples
f <- generate_f(n_samples, m, n)
# Plot histogram of the F distribution samples
hist(f, col = "blue", main = "Histogram of F distribution", xlab = "Value", ylab = "Frequency")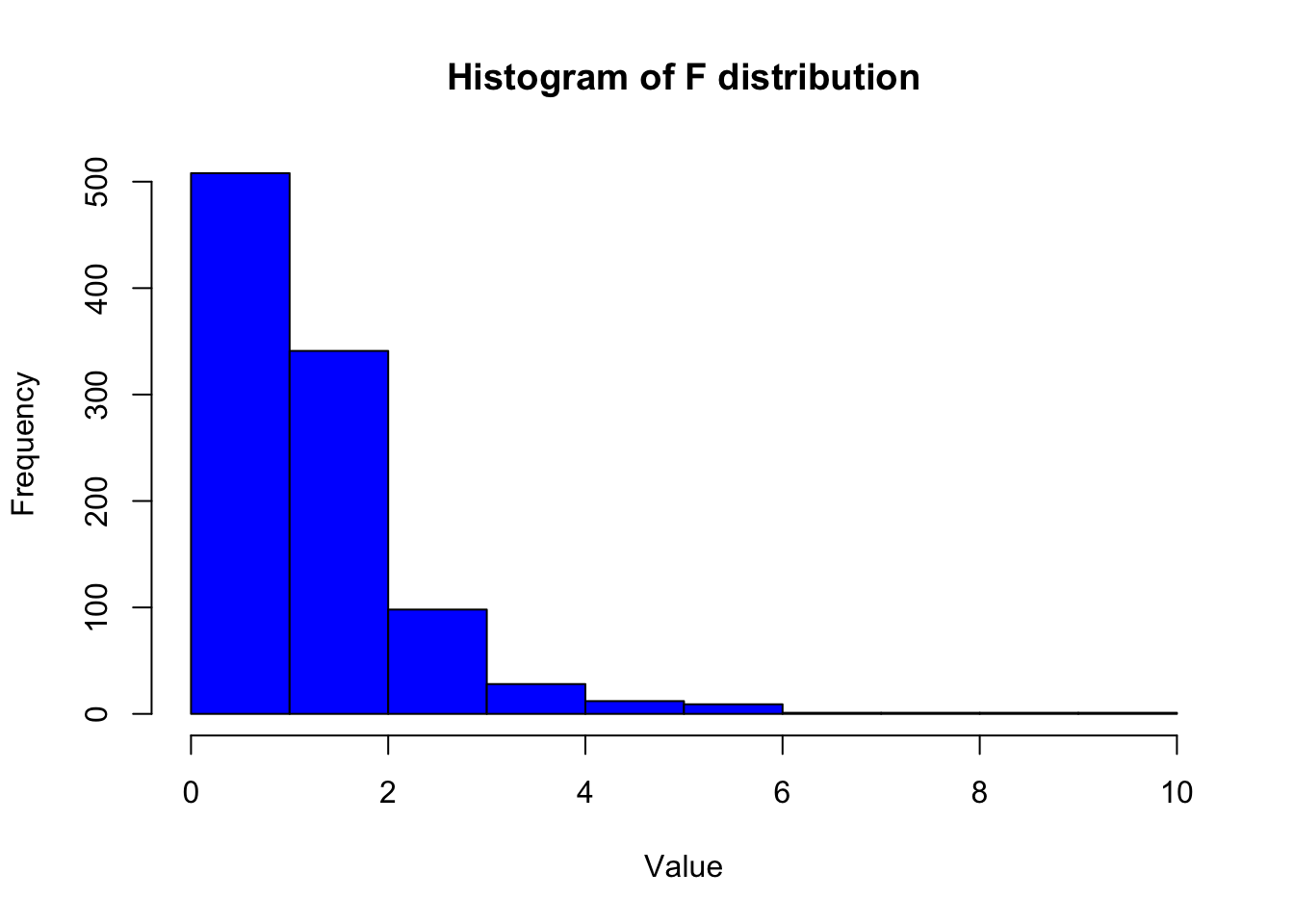
qf and ppoints# Generate theoretical quantiles from the F distribution
theoretical_quantiles <- qf(ppoints(n_samples), m, n)
# QQ plot of the theoretical quantiles against the generated samples
qqplot(theoretical_quantiles, f, main = "QQ Plot of F distribution", xlab = "Theoretical Quantiles", ylab = "Sample Quantiles")
# Add a QQ line
qqline(f, distribution = function(p) qf(p, m, n))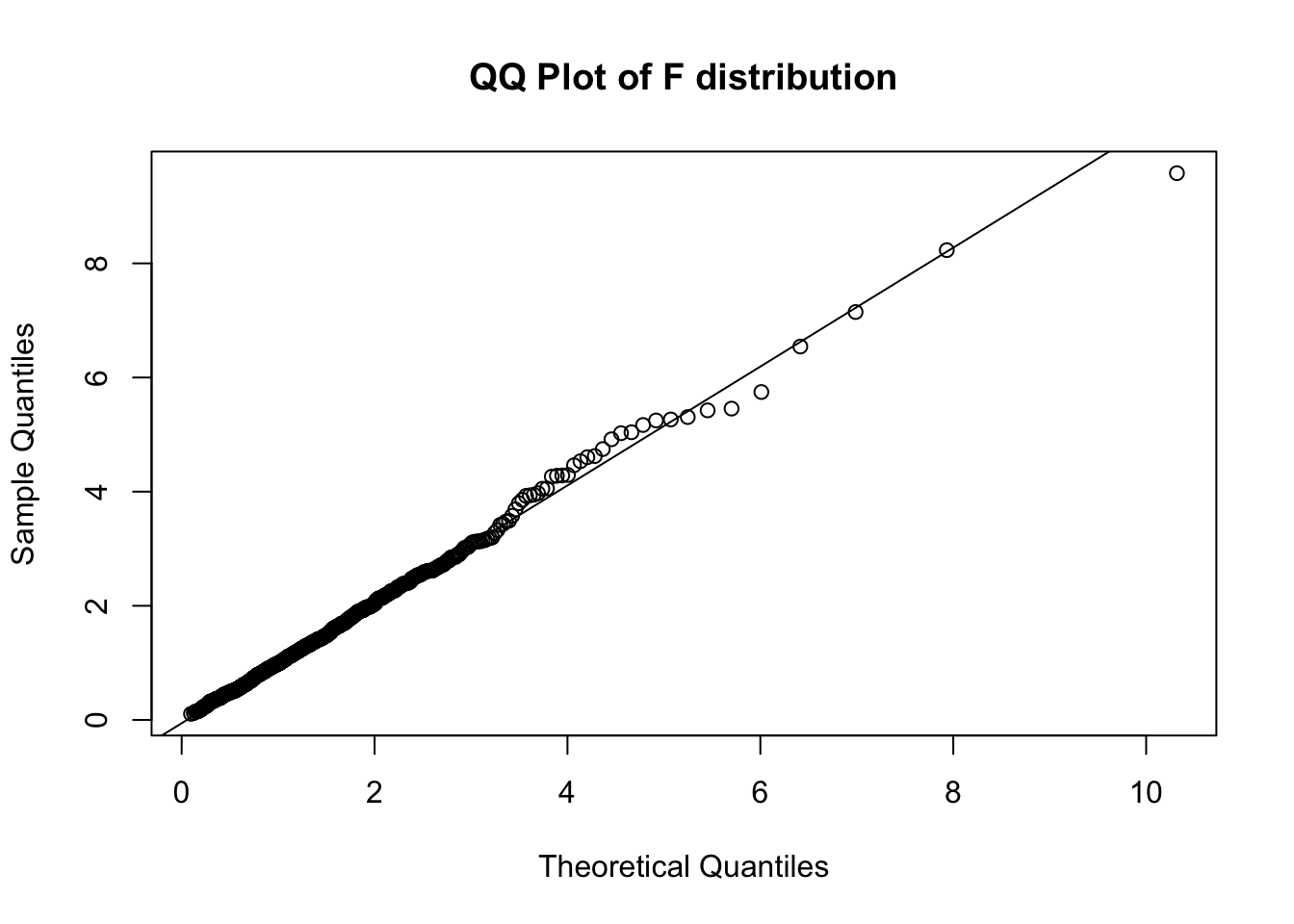
# Install invgamma package if not already installed
# install.packages('invgamma')
library(invgamma)
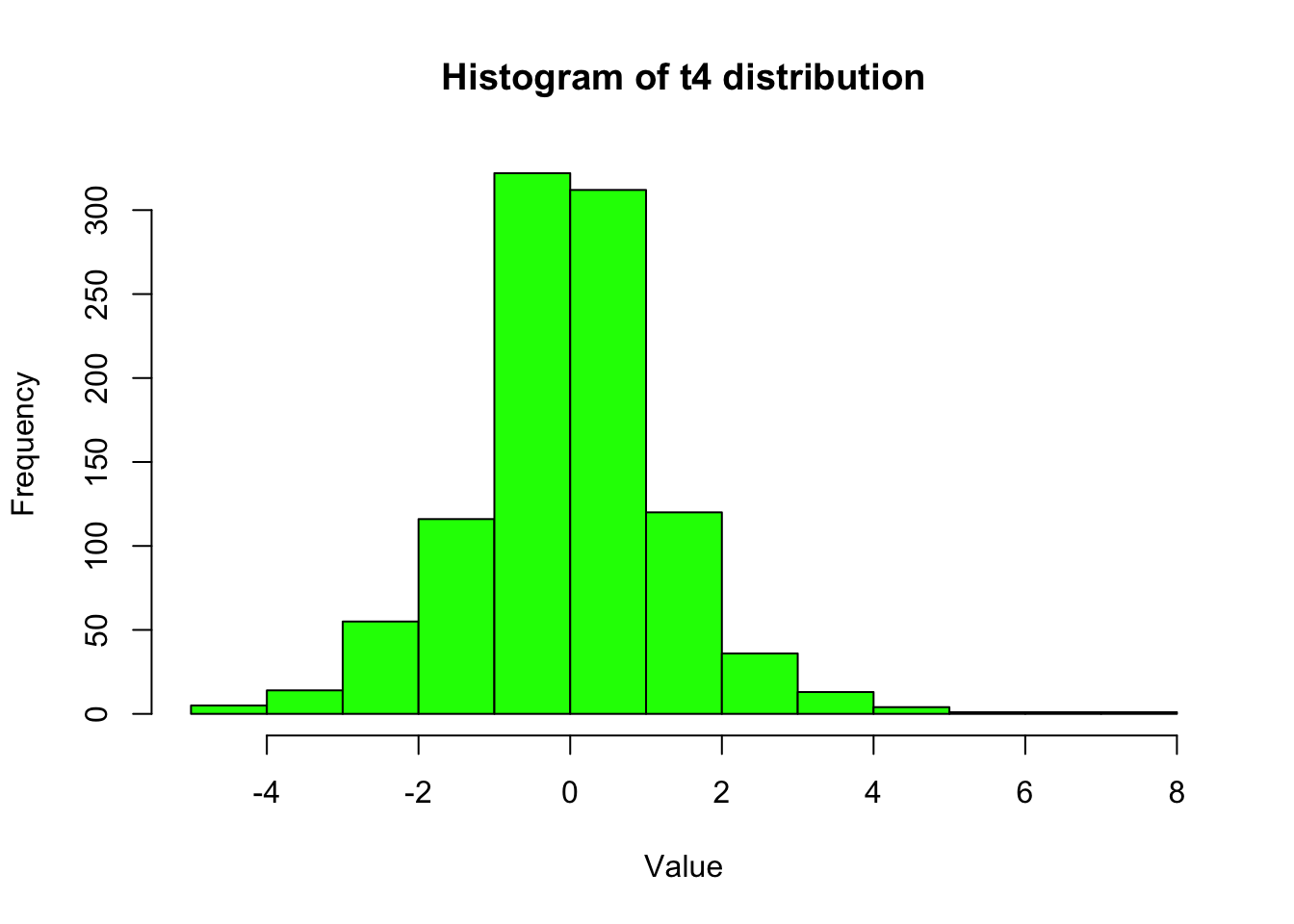
n <- 1000
v <- 4
# Generate inverse gamma random variables
vars <- rinvgamma(n, v/2, v/2)
# Generate normal random variables with variances from the inverse gamma distribution
x <- rnorm(n, 0, sqrt(vars))
# Plot histogram of the t4 distribution samples
hist(x, col = "green", main = "Histogram of t4 distribution", xlab = "Value", ylab = "Frequency")